Generic storage for heterogeneous data
by Ketil Malde; October 1, 2013
There are many options and initiatives to provide infrastructure for storing data. Most of them tend to focus on a single or very few data types, or a set of data types over a common theme. This allows a lot of structure to be added, and tight integration between data and applications. The downside is that data that don’t fit this structure is left by the roadside. Sometimes, highly structured, integrated systems are extended to take new data types into account, increasing complexity and sometimes reducing functionality or usefulness. As it is often important to retain scientific data, I think there is a need for a system that is less structured, but allows storage of more generic data types.
Framework
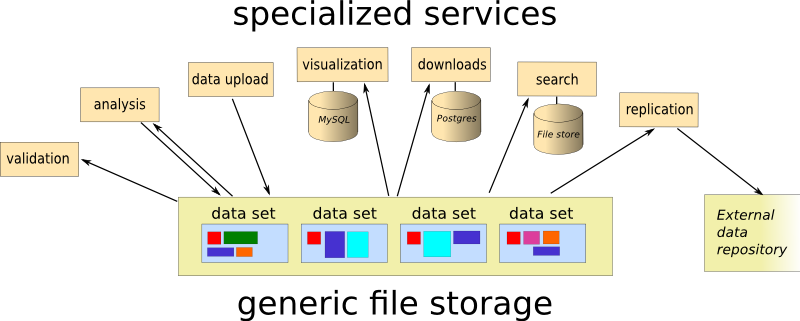
The concept is embarrassingly simple: store each data sets as a directory with the data as files, and add an XML-file called meta.xml, containing the metadata. The name of the directory must be unique, and constitutes the dataset ID. The metadata file must conform to a schema (RelaxNG, if you must know), but is mainly free text with mostly optional markup. And that’s it, really.
Use cases
So, let’s look at how this works for some use cases I think are important.
Data submission
I think perhaps the most crucial feature is ease of submission. Scientists are often required - by official policy, by publishers, or by funding agencies - to make their data publicly available, and I think most are vaguely positive to the idea. To the extent that they may make an effort of, say, five to ten minutes doing it. If it takes longer, it will get postponed, which means that it won’t happen. This is just as true if the data set contains new file types or metadata items.
Adding data is very simple: make a directory, drop some files into it, and generate the metadata. All the required bits of the metadata (e.g. file list with checksums and MIME-types) are generated automatically, and the rest is free text that may (or may not) have some markup. There are no - and I mean none - restrictions on data content, save that it must be possible to store it in a file somehow.
The incentive for the data contributor (often a scientist) is mainly that they get a receipt in the form of a unique identifier they can use to demonstrate their policy compliance.
Retrieval
The very simplest way to make data available is to point an Apache web server to the root of the data set directories. This makes it possible to browse data sets, and to download individual files. You can of course automate the download of whole data sets using standard tools like wget or curl.
Once downloaded, you can run a validation script, this will check that you have all the bits, and that they are consistent. Similarly, it is easy to set up synchronization to a remote site.
Anything more advanced will have to be implemented as a separate service, and currently there is a service that allows extraction of individual sequences from Fasta files, for instance. You can also set up a Thredds server to extract information from NetCDF files. Typical for services, these will be specialized to data they know how to handle (which they will find by scanning the metadata), and will ignore everything else.
Searching
Like all services, search is implemented as a self-contained isolated component. For generic searching over all data sets, I am using xapian to index the metadata files. In addition to straightforward text search, semantic elements (e.g. the contents of species:lepeophtheirus will find data sets concerning sea louse species.
A more specific search is searching in the data themselves. Currently, I have set up viroblast, which provides a convenient front end to BLAST, and allows users to search for molecular sequences by similarity. This is of course very data-specific, and the service crawls the metadata to identify relevant files, extract useful metadata (like species and sequence type) and incorporate them in the search service.
Automated analysis
Supporting automated analyses is a key goal. I currently have a hacked together evaluation pipeline for de novo genome assemblies, many of the intermediate results can be useful as data sets in themselves. A future enhancement to this would be to automatically build conforming data sets for e.g. sequence mappings between input sequences and the genome. Here, metadata could be generated automatically, providing the necessary provenance and relations between data sets.
Comparing to other systems
There is a plethora of initiatives to store and manage data. Let’s see what makes this system different.
An outegrated system
Many systems are integrated, they comprise a back-end database as well as a web front end, and provide analysis, data extraction services, visualization, and so on.
This system draws a clear line between data storage and services, making each service isolated from the rest of the system, and thus modular. If you are not happy with e.g. the search facilities provided, you can add your own search service, having access to the same underlying data. The important benefit from this is that technology choices are non-intrusive, and services can use technologies that are deemed most suitable independent of each other.
File based vs table based storage
Again in the name of simplicity, data is stored as plain files. This makes download and copying trivial, it is easy to verify contents with a checksum, avoids data conversion, and you don’t have to understand a schema to make use of the data. You will have to understand a file format, but chances are you already do.
Integrated systems typically store data in a relational database, this is often effective and efficient from an application point of view, but ties the data to a schema and to specific technology.
Postponing decisions
In general, I try to avoid making decisions that impose limitations of any kind. Instead, I try to make the system agnostic in as many respects as I can. Every decision is a fork in the road which limits the possible futures and adds complexity.
Lightweight metadata
There is a lot of talk in a lot of communities about ontologies. I remain skeptic, it seems to me that the process is intended to go something like this:
- precisely define every concept
- map all information using these concepts
The problem with this is that it starts with an infinite task, and then goes on to another infinite task. But maybe I’m too critical here - at any rate, I am not convinced it is really worth the trouble. Having sat in on discussions of how to name and classify things, I think there is what one could call a “design smell” to this:
When you find yourself having to make too many decisions upfront where you don’t easily see the consequences of each choice, you are doing it wrong.
And again, I’ll point to Clay Shirky on ontologies. What it boils down to, is that ontologies are - at best - a lot of work. Sometimes, they work reasonably well, for instance in the case of Gene Ontology. But Tim Berners Lee have talked about the semantic web for something like fifteen years, and yet we all still use Google and plain old hyperlinks.
So the approach here is instead to take a minimalist approach to metadata, and although it is possible to add semantic tags to the metadata text, I only add these when they have a clear use case. Kind of echoing the old IETF sentiment: no standardization without implementation. Here it is rather something like no definitions without application.
Using XML
The metadata is stored using XML, and, yes, I think I know what you are going to say. I’ve been rather critical of XML myself, it has grown to be a very complex standard, XML documents are no longer simple or easy to read (eliminating most of the advantages of a textual format), with a spaghetti of standards and links and namespaces and the kitchen sink. And software to read and manipulate XML tend to be complex and heavyweight.
But in my view, there are two ways of using XML:
- encoding structure as text
- adding structure to text
Since the SGML days, a lot of XML development - including e.g. datatypes and namespaces - have targeted the former, perhaps reaching a temporary apex with Microsoft’s document “standard”, several thousand pages long. Frankly, this is only marginally better than the memory dump that used to be Word’s file format.
The second alternative is what I use - the metadata is mostly free text, but allowing some semantic markup. So the user can tag certain elements to make it easier to index and search - but she isn’t required to do it, and the free text aspect is the most important. Meaning that the submitter will select what metadata to put in.
I’m still not sure that marking up keywords (or rather, key structures:) is a redeeming case for XML, but at least it buys a few things, like schema validation. Ideally, the system should be able to identify key structures from the text directly, but tagging them makes it easier (especially in the start), and perhaps more importantly, accelerates learning - if the system knows that “lepeophtheirus” mostly occurs in a species context, it can infer that the word is relevant to this context - also when it is not explicitly tagged. One obvious use for this is to aid the submitter, when she types in free text metadata, the editing software can scan the text for known keywords, and suggest possible contexts to apply. So it would say something equivalent of “I notice you typed Atlantic salmon, perhaps you mean the species Salmo salar? Click OK if this is what you want”.
No compulsory fields
I think this is crucial, lots of people want e.g. compulsory geographical location, for instance. For all data. Which probably seems to make sense if you are collecting data in the field, and usually want to plot it in a map. And the argument is that all data has some kind of location, so why not? If it’s from a lab, surely you can just enter the lab’s coordinates, no?
No! For one, it is irrelevant, and it will only annoy the submitter, who will have to look up the GPS coordinates of whatever lab he used. And it is very unlikely that somebody is going to look for lab data using a map. And by being forced to submit irrelevant data reduces confidence in the system, and the user is tempted to just enter nonsensical data, actively reducing the value of the system as well as the other data.
Even when geographical location is available, GPS coordinates may not be the best way to represent them. If I have salmon from a specific river population, the river name is important, the exact location not so much. And if GPS location is what is available, I will have to search by defining a polygon covering the river, which opens up opportunity for errors. So, no. Just no.
Separation of concerns
One important feature that is derived from modularity, is that it reduces the need for anybody to understand the whole system. A data manager is concerned with running the system as a whole, and must be able to interpret the metadata and perhaps know a little Unix and scripting, but doesn’t really need to know how individual services are set up, or, in particular, what the data actually are.
In contrast, a domain expert (i.e. scientist) must of course be able to make sense of the data, but shouldn’t have to worry much about metadata structures and formats, only the actual content as it is presented in the form of text. And of course, she doesn’t need to know anything about other data types than the ones that are relevant.
I think this is very important, biologists tend to be happier not learning XML or SQL, and there are simply too many domains and formats and data types that we can reasonably expect comparatively scarce data managers to know and understand them.
Shortcuts and trade-offs
The most important shortcut is being lazy - any decision that is not crucial to progress is postponed until it becomes so. I am trying to adhere to the worse-is-better philosophy, meaning that simplicity is the most important trait. Some specifics follow:
- datasets are write-only
Many people want to be able to modify datasets. What if a value in a table is wrong? Surely, we must be able to correct it. But no. Doing so obliterates provenance and reproduction of results: I can only verify your results if I am able to use the exact same dataset.
Some use version control for datasets, which seems like a reasonable strategy. But it ties you to technology (if you did this ten years ago, you might use CVS. Would you be happy about that choice now?), and it quickly makes things complex: do you really want to deal with e.g. branching and merging for data sets? In addition, data are often bulky and binary, two properties that aren’t terribly amenable to version control. Personally, I like the data library metaphor, and nobody ever questioned libraries on the ground that the authors must be able to continually update the content of books.
So my policy is that if you are unhappy with the dataset, you are free to submit a new one, and tag it as a replacement. Call it a second edition. Disk space is cheap, complexity is expensive.
- no user management or access control
Yet another one to simply avoid. Many users want to restrict accessiblity to data. For one, I think data should be freely available to anyone, and we generally don’t have sensitive or confidential data (e.g., privacy issues). Second, user management is often the most complicated and administration-intensive part, both technically (I once coordinated multiple sites with a common user data base in LDAP, ugh), and administratively (constant trickle of user requests, removals, lost passwords, and so on).
If this becomes really necessary, I’ll suggest you upload your data in encrypted form, and manage the key distribution and user base separately.
- targeting humans, not machines
This means that we don’t enforce ontologies and use a very lightweight XML schema. Thereby, we lose precision in the metadata, by allowing researchers to type any metadata they want, we also allow sloppiness and inconsistency. But the gain is in metadata accuracy, by letting the researcher submit what he or she thinks is important, it is more likely that the metadata are relevant, and we avoid any need to invent something to satisfy the system. Using plain text also emphasizes the receiver as a human, and encourages to submitter to think about the audience, and what they will be actually interested in knowing.
Availability
I’m not sure the implementation is ready for, uh, industrial use, but if you want to peek, it’s there. That is, at http://malde.org/~ketil/datastore. If you are interested, there’s also a poster and its abstract, presented at IMDIS recently.
comments powered by Disqus

