The effect of using different substitution matrices
by Ketil; November 8, 2012
I’ve recently written about transitive alignments, and hopefully, there will be a publication out soon. One of the reviewer suggested I should use a different substitution matrix for BLAST, and so I did.
The official NCBI Blast documentation has this to say:
Experimentation has shown that the BLOSUM-62 matrix [4] is among the best for detecting most weak protein similarities. For particularly long and weak alignments, the BLOSUM-45 matrix may prove superior.
The number of a BLOSUM matrix refers to the average sequence similarity, so clearly BLOSUM-45 is more appropriate for aligning sequences that are more distantly related. I generated an updated plot that includes all the different alternatives:
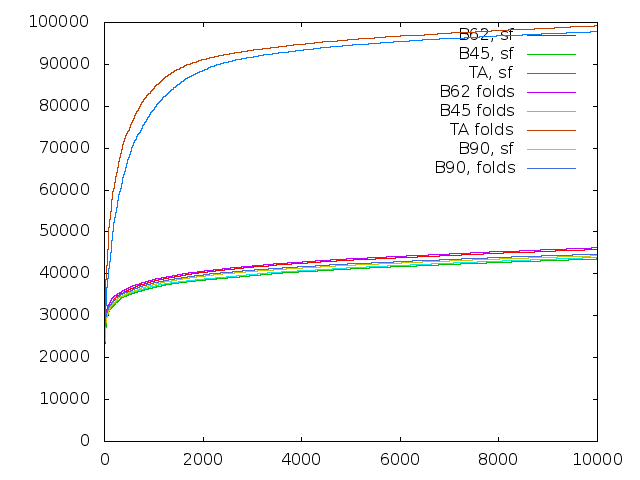
Comparing Transitive Alignments with BLAST using different substitution matrices.
Here, Bnn means using BLAST with a BLOSUM-nn matrix, and TA is of course transitive alignments. False positives along the x-axis, and true positives along they y-axis, as before - in other words, higher faster is better. We make the following observations:
- there is very litte effect at all from varying the substitution matrix
- BLOSUM-45 is actually a bit worse than BLOSUM-62.
- Perhaps less surprising, so is BLOSUM-90.
- Transitive alignments remains quite a bit better.
(Interestingly, a bug in the software used to calculate BLOSUM-62 not only makes it inaccurate, but also happens to improve its performance. This could perhaps explain the apparent effectiveness of BLOSUM-62.)
comments powered by Disqus

